Secondary Structure Prediction¶
- We will use one of the differentially expressed in tomato pollen transcriptome under head stress.
I have retrive the amino acid sequences for Solyc06g050510 from SolGen website.
MKRHIHYNAHPIDPHPFEAFWYGSWQAVERLRINMGTITTHVLVDGEVIEENIPVTNLRMRSRKATLSDCAC FLRPGLEVCVLSIPYQGENSGDEKDVKPVWIDGKIRSIERKPHELTCTCKFHVSVYVTQGPPPILKKTLSKE IKMLPIDQIAVLQKLEPKPCENKRYRWSSSEDCNSLQTFKLFIGKFSSDLTWLMTASVLKEATFDVRSIHNQ IVYEIVDDDLVRKETNSNQHSYSVNFKLEGGVQTTTVIQFNRDIPDINSTSDLSESGPLVLYDLMGPRRSKR RFVQPERYYGCDDDMAEFDVEMTRLVGGRRKVEYEELPLALSIQADHAYRTGEIEEISSSYKRELFGGNIRS HEKRSSESSSGWRNALKSDVNKLADKKSVTADRQHQLAIVPLHPPSGTGLTVHEQVPLDVDVPEHLSAEIGE IVSRYIHFNSSSTSHDRKASKMNFTKPEARRWGQVKISKLKFMGLDRRGGTLGSHKKYKRNTTKKDSIYDIR SFKKGSVAANVYKELIRRCMANIDATLNKEQPPIIDQWKEFQSTKSSQRESGDHLAMNRDEEVSEIDMLWKE MELALASCYLLDDSEDSHAQYASNVRIGAEIRGEVCRHDYRLNEEIGIICRLCGFVSTEIKDVPPPFMPSSN HNSSKEQRTEEATDHKQDDDGLDTLSIPVSSRAPSSSGGGEGNVWALIPDLGNKLRVHQKRAFEFLWKNIAG SIVPAEMQPESKERGGCVISHTPGAGKTLLIISFLVSYLKLFPGSRPLVLAPKTTLYTWYKEVLKWKIPVPV YQIHGGQTFKGEVLREKVKLCPGLPRNQDVMHVLDCLEKMQMWLSQPSVLLMGYTSFLTLTREDSPYAHRKY MAQVLRQCGLLILDEGHNPRSTKSRLRKGLMKVNTRLRILLSGTLFQNNFGEYFNTLTLARPTFVDEVLKEL DPKYKNKNKGASRFSLENRARKMFIDKISTVIDSDIPKKRKEGLNILKKLTGGFIDVHDGGTSDNLPGLQCY TLMMKSTTLQQEILVKLQNQRPIYKGFPLELELLITLGAIHPWLIRTTACSSQYFKEEELEALQKFKFDLKL GSKVKFVMSLIPRCLLRREKVLIFCHNIAPINLFLEIFERFYGWRKGIEVLVLQGDIELFQRGRIMDLFEEP GGPSKVMLASITTCAEGISLTAASRVILLDSEWNPSKSKQAIARAFRPGQDKVVYVYQLLATGTLEEEKYKR TTWKEWVSSMIFSEDLVEDPSHWQAPKIEDELLREIVEEDRATLFHAIMKNEKASNMGSLQE
Copy and paste the amino acid sequence in the box and label the sequence.
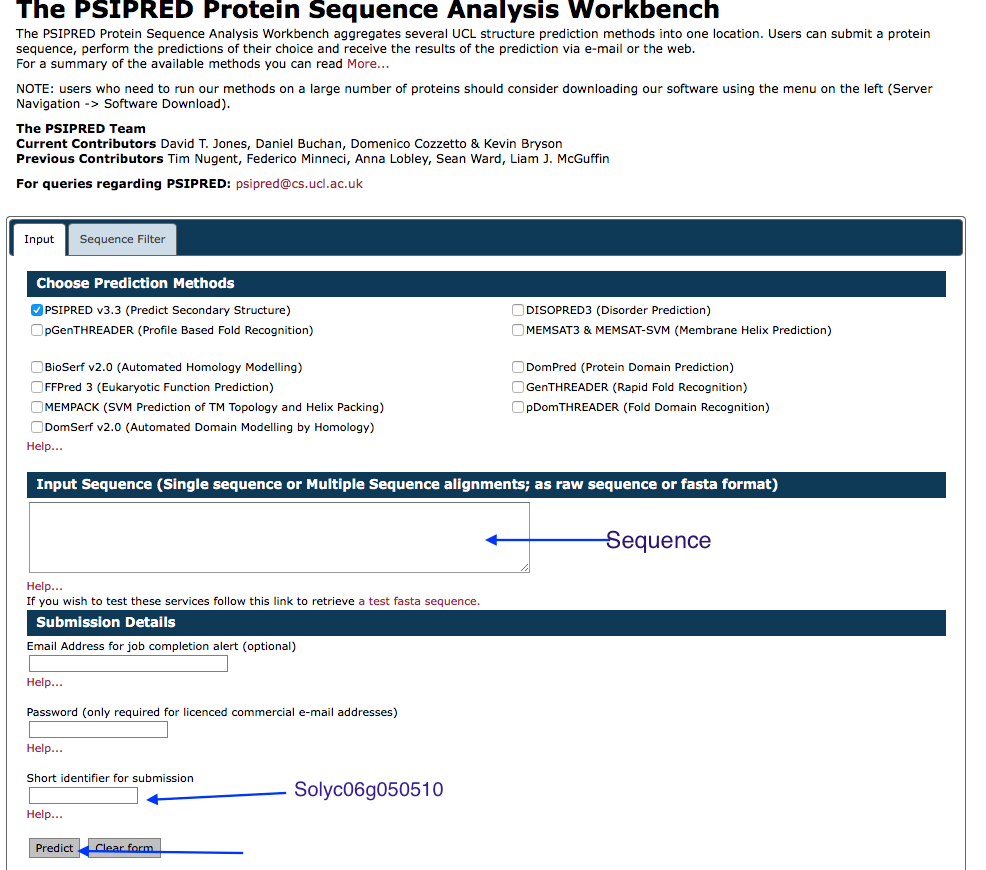
- We will use the previous submitted results:
http://bioinf.cs.ucl.ac.uk/psipred/result/e3f48c8e-28ff-11e7-879a-00163e110593
Tertiary Structure Prediction¶
Frist find a structure similar to above sequence in PBD. We will use DELTA BLAST to search PBD.
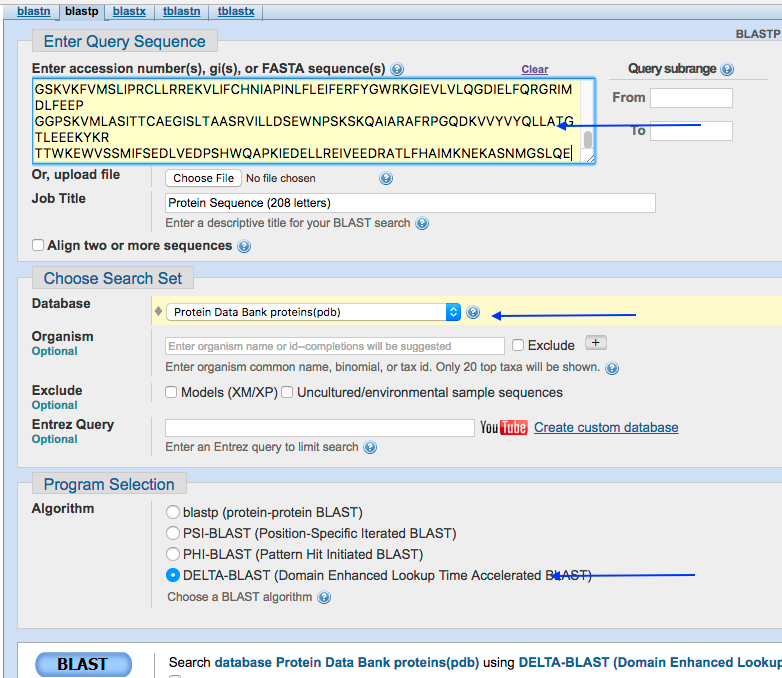
2. Click on the first significant hit to access the PDB. In case, you don’t have BLAST results, use the following link to access the previous results. Link
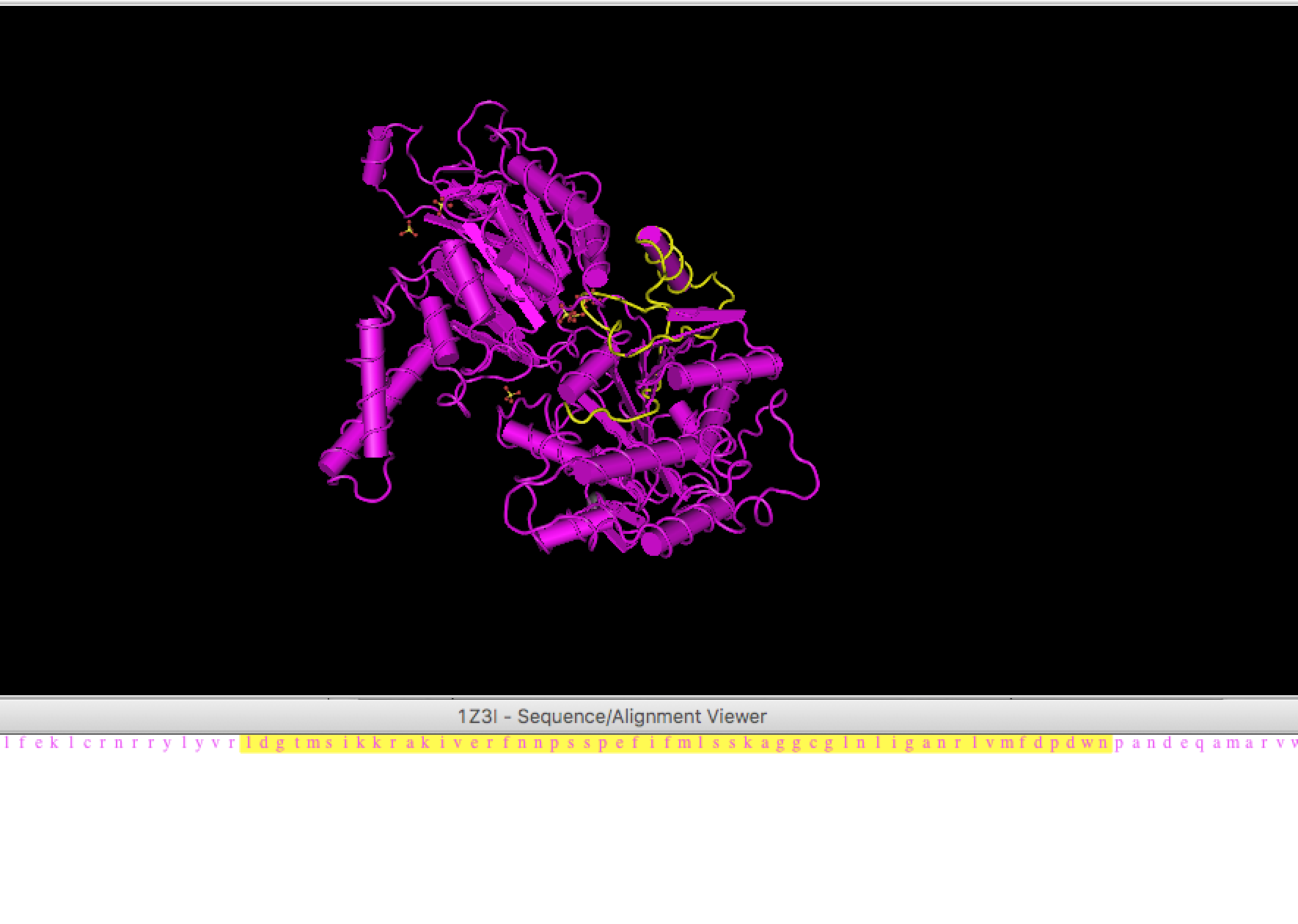
- RCSB provides curated content of PDB and use PDB ID: 1Z3I to visualize the protein in RCSB.
4. Perform a multiple sequence alignment to find conserved sequences. |
:a:. Retrive sequence from databank
https://github.com/ajwije/2017_spring_Bioinfo_class/blob/master/rad54.fasta
:c:. Use Tcoffee server and align the sequences using structural information:
You can download crystal structure information from PDB in Cn3 format.
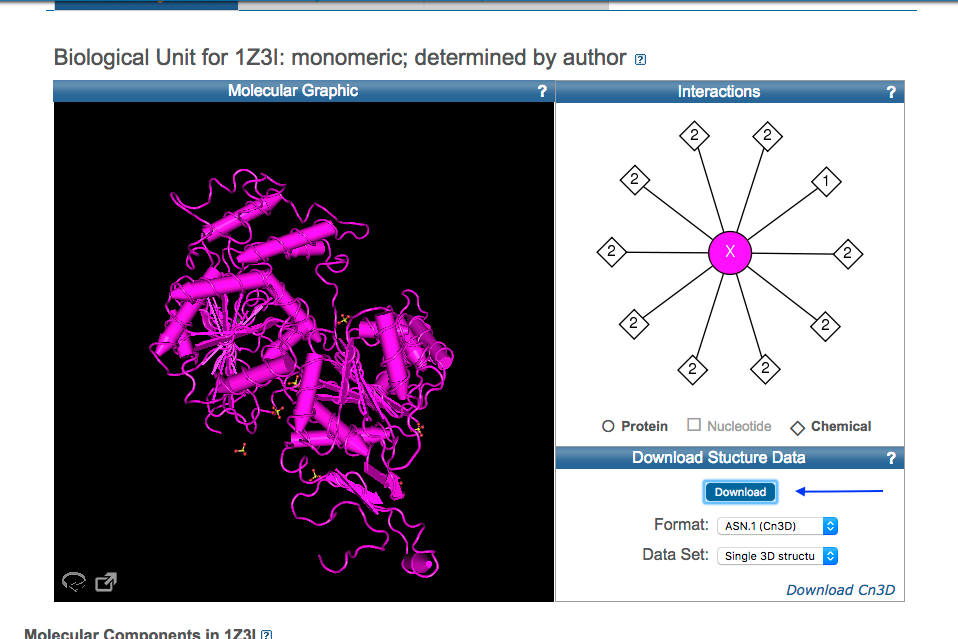
Download Cn3D software from NCBI and install it on your computer.
Open above Cn3 file using the Cn3D software.
Go to sequence viewer
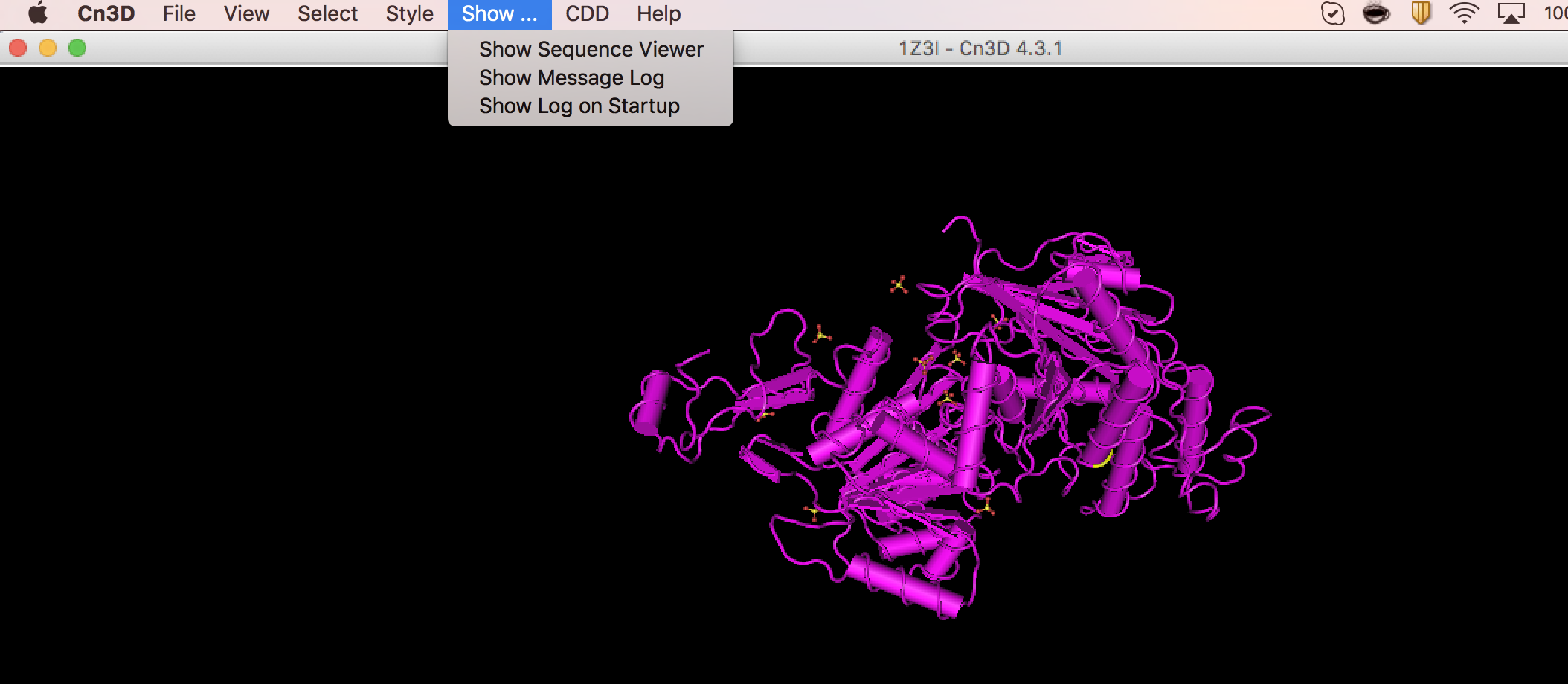
Under view, select find pattern:
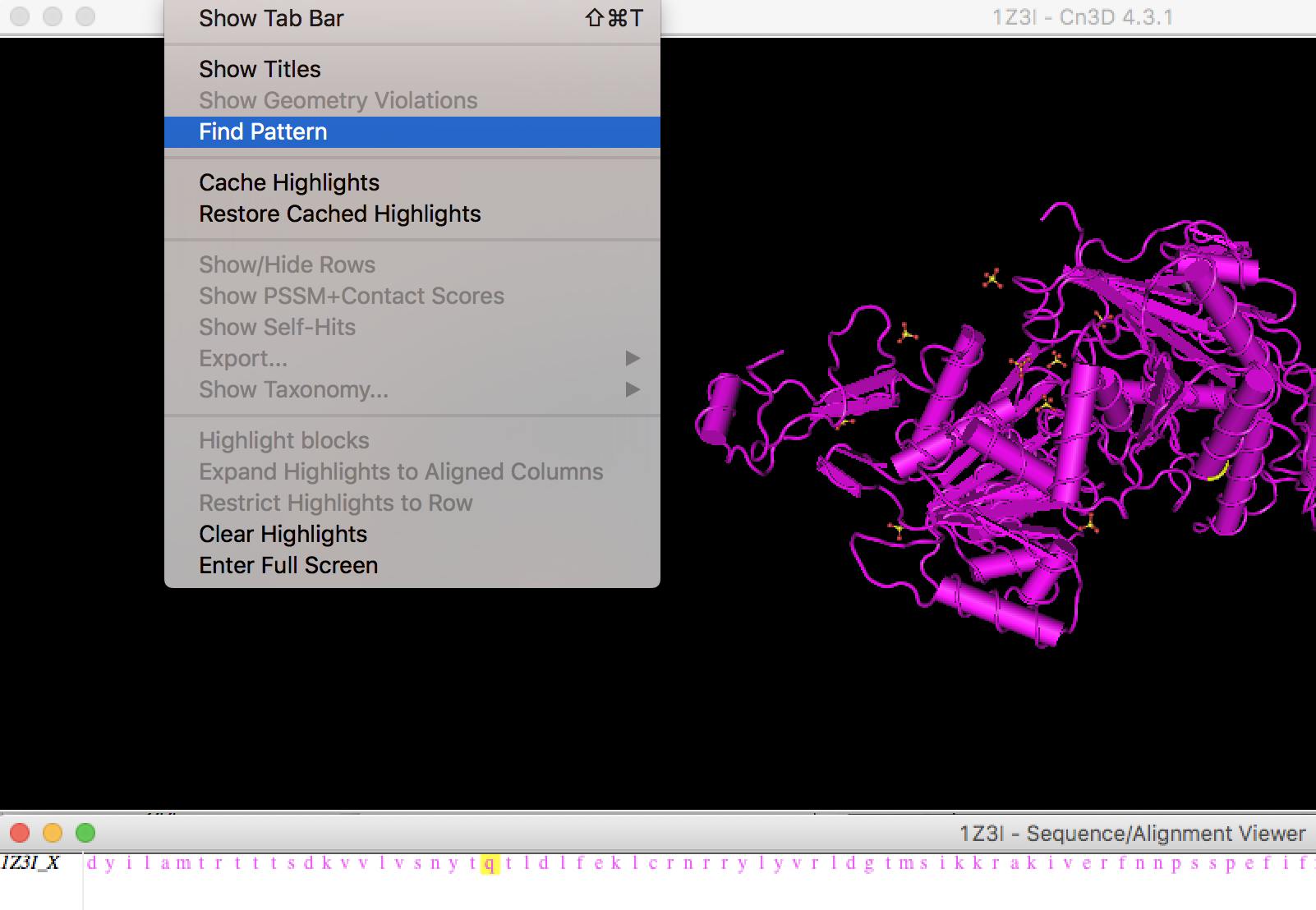
Copy a conserved region from multiple sequence alignment in the search window and click OK:

You will see conserved region displayed on the crystal structure.